
Continuing advances in the acquisition and processing of three-dimensional data has made the use of computed tomography (CT) scanning an essential tool in paleontology, entomology, geology, and other biodiversity disciplines in which the visualization of surface and internal structures of 3D objects is critical. IDigBio was delighted to collaborate with staff at the High-resolution X-ray CT Facility (UTCT) at the University of Texas’ Jackson School for Geosciences the week of 23-25 February to offer a CT scanning short course focused on the utilization of software for visualizing and analyzing CT data. Thanks to Jessie Maisano and UTCT team members Matt Colbert, David Edey, Gary Zuker, and Travis Clow, short course attendees were treated to three full days of well-received training with plenty of time for individualized application to personally significant research data.
 Ten participants, including graduate students and early career professionals from throughout the United States, traveled to Austin for the event. Perquisites for attendance included immediate need for applying CT processing skills to on-going research, availability of Avizo visualization software at one’s home institution, and availability of one or more existing 3D datasets or physical objects to be scanned at UTCT. About 30 applicants were considered for the limited space, all of which met the criteria, making participant selections challenging. Limited acceptance ensured one participant per station in the ten-station lab and provided for optimal hands-on instruction.
Ten participants, including graduate students and early career professionals from throughout the United States, traveled to Austin for the event. Perquisites for attendance included immediate need for applying CT processing skills to on-going research, availability of Avizo visualization software at one’s home institution, and availability of one or more existing 3D datasets or physical objects to be scanned at UTCT. About 30 applicants were considered for the limited space, all of which met the criteria, making participant selections challenging. Limited acceptance ensured one participant per station in the ten-station lab and provided for optimal hands-on instruction.
Following a first-day introduction to iDigBio by Gil Nelson, the UTCT team launched immediately into course content, with an introduction to computed tomography and its varied uses across a variety of disciplines, followed by a tour of the UTCT facility. Maisano demonstrated how objects are readied and mounted for scanning and the intricacies of the Zeiss (formerly Xradia) MicroXCT scanner. Colbert offered an overview of the larger and completely upgraded North Star scanner, its output and idiosyncrasies, and discussed limitations and possibilities with the technology.
immediately into course content, with an introduction to computed tomography and its varied uses across a variety of disciplines, followed by a tour of the UTCT facility. Maisano demonstrated how objects are readied and mounted for scanning and the intricacies of the Zeiss (formerly Xradia) MicroXCT scanner. Colbert offered an overview of the larger and completely upgraded North Star scanner, its output and idiosyncrasies, and discussed limitations and possibilities with the technology.
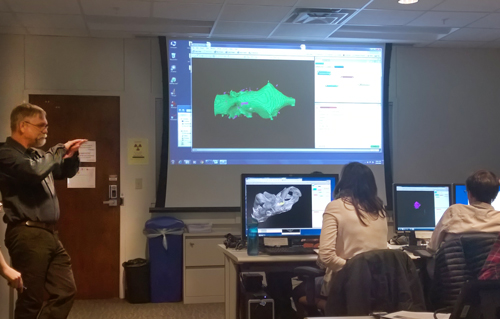 With a better understanding of CT data acquisition as a background, the afternoon session began intensive training in the use of two visualization, analysis, and processing software packages. Participants first loaded a demonstration dataset to explore ImageJ, a free, open source, sophisticated, extensible image processing software designed for processing scientific multidimensional images and suitable for large CT datasets. ImageJ training was followed by a focus on Avizo, among the most popular, powerful, and widely used 3D data manipulation and processing applications. Participants also learned to use QuickTime to animate their output.
With a better understanding of CT data acquisition as a background, the afternoon session began intensive training in the use of two visualization, analysis, and processing software packages. Participants first loaded a demonstration dataset to explore ImageJ, a free, open source, sophisticated, extensible image processing software designed for processing scientific multidimensional images and suitable for large CT datasets. ImageJ training was followed by a focus on Avizo, among the most popular, powerful, and widely used 3D data manipulation and processing applications. Participants also learned to use QuickTime to animate their output.
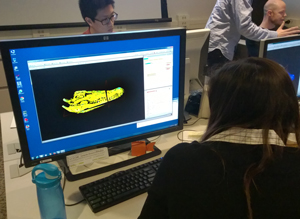 Attendees used a sample crocodile skull dataset in conjunction with direct instruction and an excellent tutorial produced by the UTCT staff to explore the basics of Avizo before gradually making the transition to their own research datasets. Most of Tuesday afternoon and virtually all of Wednesday were structured to allow participants to work with their personal datasets and research goals, with hands-on assistance from lab staff. The opportunity to receive personalized tips and assistance from lab instructors as well as other participants was invaluable and was among the most successful components of the short course. Participants developed important and potentially long-lasting relationships with each other as well as lab staff, availed themselves of every opportunity to seek assistance from short course instructors, and worked diligently up to the short course’s final bell at 5:00 p.m. on the 25th.
Attendees used a sample crocodile skull dataset in conjunction with direct instruction and an excellent tutorial produced by the UTCT staff to explore the basics of Avizo before gradually making the transition to their own research datasets. Most of Tuesday afternoon and virtually all of Wednesday were structured to allow participants to work with their personal datasets and research goals, with hands-on assistance from lab staff. The opportunity to receive personalized tips and assistance from lab instructors as well as other participants was invaluable and was among the most successful components of the short course. Participants developed important and potentially long-lasting relationships with each other as well as lab staff, availed themselves of every opportunity to seek assistance from short course instructors, and worked diligently up to the short course’s final bell at 5:00 p.m. on the 25th.







