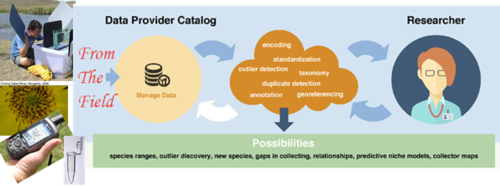
Field to Database
| Field to Database | |
|---|---|
| Quick Links for Field to Database | |
| Link to Agenda | |
| Workshop Presentation Biblio Entries | |
| Workshop Blog | |
Apply Now
Spots are limited. Application Form is live. Apply now to save your spot! First-come, first-served.
General Information
This workshop's aim is to investigate current trends in collecting, and focus on best practices and skills development for supporting the collection and sharing of robust, fit-for-research-use data. This 4-day short course is designed to be hands-on and will mix lectures with field work and participant exercises and presentations.
Planning Team
Deb Paul (iDigBio), Katja Seltmann (TTD-TCN, MNH), François Michonneau (FLMNH - iDigBio), Derek Masaki (USGS - BISON), Pam Soltis (FLMNH - iDigBio PI), Shari Ellis (iDigBio), Kevin Love (iDigBio)
About
Skill Level
Some exposure to R is required. This workshop expects you have some experience with R. If you are new-ish to R, we request you take an intro to R course before the workshop. There are several good options:
- Try R (Code School course)
- intro to R(Coursera course starts Feb 2nd).
- Beginner Course: Up and Running with R with Barton Poulson (course at lynda.com)
- Intermediate Course: R Statistics Essential Training with Barton Poulson(course at lynda.com)
Instructors: François Michonneau (FLMNH - iDigBio), Katja Seltmann (TTD-TCN, AMNH), Derek Masaki (USGS), Matt Collins (ACIS - iDigBio)
Assistants: Deborah Paul (FSU - iDigBio), Matt Cannister (USGS)
Who: The course is aimed at graduate students, postdocs, research staff, and other researchers.
Where: iDigBio in Gainesville, FL
Requirements:
- Participants must bring a laptop with a few specific software packages installed.
- Participants must have some knowledge of R. This is not a beginner-level course. There are introductions to R you can take on-your-own before the workshop.
- If you will be traveling from out of town, you will need to make your own travel arrangements.
Contact: Please email Deb Paul, dpaul@fsu.edu for questions and information not covered here.
Twitter: #field2db
Tuition for the course is free, but prior registration is required for attending. You can register here.
Software Installation Requirements
Software needed for Field to Database Course at iDigBio
Mac OS X
- Text Editor
- We recommend Text Wrangler. In a pinch, you can use nano, which should be pre-installed.
- RStudio + R
- Install R by downloading and running this .pkg file from CRAN. Also, please install the RStudio IDE.
- Spreadsheet
- If you already have a spreadsheet program installed, like LibreOffice, Excel or OpenOffice, you can use whatever you already have. If you don't have a spreadsheet program, please download and install LibreOffice from http://www.libreoffice.org/download/libreoffice-fresh/
PC
- Text Editor
- Notepad++ is a popular free code editor for Windows. Be aware that you must add its installation directory to your system path in order to launch it from the command line (or have other tools like Git launch it for you). The instructions to modify your path are available online here. Please ask your instructor to help you do this.
- RStudio + R
- Install R by downloading and running this .pkg file from CRAN. Also, please install the RStudio IDE.
- Spreadsheet
- If you already have a spreadsheet program installed, like LibreOffice, Excel or OpenOffice, you can use whatever you already have. If you don't have a spreadsheet program, please download and install LibreOffice from http://www.libreoffice.org/download/libreoffice-fresh/
Linux
- Text Editor
- Kate is one option for Linux users. In a pinch, you can use nano, which should be pre-installed.
- RStudio + R
- You can download the binary files for your distribution from CRAN. Or you can use your package manager, e.g. for Debian/Ubuntu run
apt-get install r-base. Also, please install the RStudio IDE.
- You can download the binary files for your distribution from CRAN. Or you can use your package manager, e.g. for Debian/Ubuntu run
- Spreadsheet
- If you already have a spreadsheet program installed, like LibreOffice, Excel or OpenOffice, you can use whatever you already have. If you don't have a spreadsheet program, please download and install LibreOffice from http://www.libreoffice.org/download/libreoffice-fresh/
- You must RSVP that the required software is installed, prior to the workshop. Instructors are available to help - see your email for their contact information.
We use Adobe Connect extensively in this workshop. Please perform the systems test using the link below. Also, you will also need to install the Adobe Connect Add-In to participate in the workshop.
Goals
- Investigate, observe, discover leading-edge trends in field collecting.
- Provide examples of best practices for data collecting and data sharing including such data as field data, identifiers, trait data, and environmental variables.
- Explore data tools, to include software such as R, but also field apps.
- Convey the concept of, importance, and methods for how to create reproducible research workflows.
- Illustrate how data gets from the field into a collection database and into an aggregator's database.
- Discuss how data gets published and discovered.
Objectives
- Students participate in field collecting with subject-matter experts and present what changes they plan to make to their collecting practices in a workshop presentation.
- Subject-matter experts share what they have learned from seeing / talking with others on this topic.
- Students work through examples to demonstrate mastery of skills for transforming, enhancing, standardizing data.
- Through comments, discussion, and perhaps post-workshop survey, students demonstrate they grasp the importance of metadata and understand the conceptual difference between data and metadata.
- Students write a post-workshop blog post, prepare a report, or presentation, to synthesize what was learned and pay-it-forward.
Our curriculum overview
- Day 1: Why a Field-to-Database Biodiversity Informatics Workshop? On Site Field Demos from Invited Experts from Paleontology, Ornithology, Ecology, Marine Science, Entomology, and Botany
- Day 2: Student 3-minute presentations. General issues in field data collection to data synthesis. Getting started with R.
- Day 3: Data exploration using R. Import and display. From raw data to technically correct data. From technically correct data to consistent data. File output. Writing processed data to file.
- Day 4: Using R to access biodiversity APIs. Publishing data on iDigBio. Publishing data on DataDryad. Review, Wrap-up, Survey, Next Steps.
The concepts, skills, and tools we teach are domain-independent, but example problem cases and datasets will be taken from organismal and evolutionary biology, biodiversity science, ecology, and environmental science.
Updates to course wiki will be posted to this website as they become available.
Workshop Evaluation
- link to pre-workshop survey (if we do one)
- Post Workshop Survey Results
Agenda
- AdobeConnect #field2database Room (if we are going to use).
- Pre-workshop meeting and dinner at Piesano's, 6 PM Sunday March 8th, 2015. Piesano's is at NW 13th St. and 1250 W. University Ave. in Gainesville. All are welcome. Please do RSVP to Deb Paul, dpaul@fsu.edu
| Course Overview - Day 1, Monday March 9th | ||
|---|---|---|
| 8:30-9:00 | Registration. name tags, wired/wireless, adobeconnect, check-in. |
All, Deb Paul (iDigBio) |
| 9:00-9:20 | Welcome and Introduction to iDigBio | Deb Paul (iDigBio), Pam Soltis (iDigBio PI) |
| 9:20-10:00 | Why a Field-to-Database Biodiversity Informatics Workshop? | Katja Seltmann (AMNH), Pam Soltis (iDigBio PI) and Charlotte Germain-Aubrey (iDigBio Post Doc) |
| 10:00-10:15 | Break | |
| 10:15-11:30 | Invited Speaker Presentations From the Field (10 to 15 min each) |
Katja Seltman (Lead); Emilio Bruna; Justin Woods; Mike Webster; Andrew Short; Grant Godden; François Michonneau (or Gustav Paulay) |
| 11:30 - 12:00 | Transport to Natural Teaching Area | (vans) |
| 12:00 - 1:00 | Lunch (Brown Bag provided) | (organizers set up demo areas) |
| 1:00-4:00 | On Site Field Demos from Invited Experts | Katja Seltmann (Lead) |
| 4:00-5:00 | Return to 105 for Wrap-up and Homework: Create a 3 min presentation. | Katja Seltmann (Lead) |
| 6:00 | Dinner on your own. | Potential to have dinners together if desired. |
| Course Overview - Day 2, Tuesday March 10th | ||
| 8:30-9:00 | Check in, answer questions | All, Deb Paul |
| 9:00-11:00 | Discussion and Homework Presentations. (3 min. each participant) |
Katja Seltmann and Invited Experts (Lead) |
| 11:00-11:30 | General issues in field data collection to data synthesis. | Derek Masaki and Katja Seltmann (Lead) |
| 11:30-12:30 | Reproducible Research | Derek Masaki (Lead) |
| 12:30-1:30 | Lunch | (on your own, or provided) |
| 1:30-1:45 | Review of data set: identify issues, errors. | Derek Masaki (Lead) |
| 1:45-5:00 | Getting started with R | François Michonneau (Lead) |
| 5:00-5:30 | Review / Homework? / Preview of tomorrow | |
| Course Overview - Day 3, Wednesday March 11th | ||
| 8:30-9:00 | Check in, answer questions | All, Deb Paul |
| 9:00-10:00 | Data exploration using R. Import and display. | Derek Masaki (Lead) |
| 10:00-12:00 | From raw data to technically correct data. | Derek Masaki (Lead) |
| 12:00-1:00 | Lunch | on your own (or provided) |
| 1:00-2:00 | From technically correct data to consistent data. | Derek Masaki (Lead) |
| 2:00-3:00 | File output. Writing processed data to file. | Derek Masaki (Lead) |
| 3:00-4:30 | ||
| 5:00-5:00 | Review / Wrap-up / Preview of tomorrow | |
| Course Overview - Day 4, Thursday March 12th | ||
| 8:30-9:00 | Check in, answer questions | All, Deb Paul |
| 9:00-12:00 | Using R to access biodiversity APIs | Francois Michonneau, Matt Collins (Leads) |
| 12:00-1:00 | Lunch | on your own (or provided) |
| 1:00-1:45 | Publishing data on iDigBio | Molly Phillips, Matt Collins (Leads) |
| 2:30-4:00 | Publishing data on DataDryad (includes discussion of metadata) | Todd Vision, DataDryad (Lead) |
| 4:00-5:00 | Review, Wrap-up, Survey, Next Steps. | 1 slide lightning talks by volunteer participants: One thing learned, one thing they realize they need to learn more about -- their plans for learning more, and how they plan to pay-it-forward (to share what they’ve learned with colleagues / fellow students -- through talks? / blog posts? / seminars?) |
| Optional Evening Session -- on working with their own data? | ||
Future plans: Scaling it up: Demo using the iPlant Discovery Environment (DE)
Link to Workshop Report
Logistics
- Logistics & Hotel Information (for any out-of-towners)
- Where to find food
- Workshop Calendar Announcement
- Participant List
Adobe Connect Access
Adobe Connect will be used to provide communication between all present at the workshop. Remote participants will be able to listen to lecture portions only.
We use Adobe Connect extensively in this workshop. Please perform the systems test using the link below. Also, you will also need to install the Adobe Connect Add-In to participate in the workshop. Adobe Connect Systems Test
Presentation Documents and Links
- SQL commands list
- SHELL commands list
- R lesson
- Getting started with Open Refine
- Google's R Style Guide
- make code easier for you, and others, to understand
Biodiversity APIs
- taxize tutorial
- taxize on github
- ridigbio
- Open Tree of Life APIs
- Introduction to the VertNet API
- rgbif on github
- rgbif tutorial
- rgbif: Interface to the Global Biodiversity Information Facility API
Workshop Recordings
Day 1
- 9:00am-10:00am
- 10:15am-11:30pm
- 4:00pm-5:00pm
Day2
- 9:00am-11:00am
- 11:00am-11:30pm
- 11:30pm-12:30pm
- 1:30pm-1:45pm
- 1:45-5:00pm
- 5:00-5:30pm
Day3
- 9:00-10:00
- 10:00-12:00
- 1:00-2:00
- 2:00-3:00
Day4
- 9:00-12:00
- 1:00-2:30
- 2:30-4:00
- 4:00-5:00
Related Workshop Resources and Links
- Data Carpentry Materials on GitHub
- Ten Simple Rules for the Care and Feeding of Scientific Data. Goodman et al
- Code and Data for the Social Sciences: A Practitioner's Guide. Matthew Gentzkow, Jesse M. Shapiro Chicago Booth and NBER March 10,2014
- Nine simple ways to make it easier to (re)use your data. White et al.
- You want to learn SQL independently? Try Head First SQL
- Head First Excel, O'Reilly
- Check out DataONE
- They've got a great Software Tools Catalog
- Put standard metatdata with your data. Wondering how to do that? Check out DataONE's Morpho Tool available under the tools menu at https://knb.ecoinformatics.org/.
- Why? Makes your data re-useable, and better still, makes your data discoverable. Get cited for your datasets in addition to your published papers!
- Making Sense of Data Free online course at Google.
- " Do you work with surveys, demographic information, evaluation data, test scores or observation data? What questions are you looking to answer, and what story are you trying to tell with your data? This self-paced, online course is intended for anyone who wants to learn more about how to structure, visualize, and manipulate data. This includes students, educators, researchers, journalists, and small business owners."
- Using Open Refine? Want to compare your taxon names against a standard list? Try this reconciliation service.
- Read Gaurav ' Blog post first: http://gbif.blogspot.com/2013/07/validating-scientific-names-with.html
- Then, give it a try. The google plus Open Refine community will help you figure it out (it's not hard).
Links from You
- How about you? Got a favorite resource - a book?, a website? to share with your classmates?
- Data Science at the Command Line
- Free Training Resources for UF students, faculty, and staff UF provides free access to over 2600 online training courses through Lynda.com. Does your institution have similar free training opportunities?
Related Blog Posts and Photos
- Inaugural Data Carpentry Workshop by Tracy K. Teal
- Our First Data Carpentry Workshop by Karen Cranston
- Tales from the First Data Carpentry Workshop by Deb Paul, May 2014
- Data Carpentry, Please can we have some more?! by Deb Paul, 15 Oct 2014
- Data Carpentry Facebook Photo Album